
| Index to this page |
Here is an overview.

This page examines the first step:
| Quality control. Occasionally RNA polymerase will select and insert an incorrect, mismatched, ribonucleotide. When this occurs in bacteria (and perhaps in all organisms), the enzyme backs up, removes the incorrect nucleotide (and the one preceding it) and tries again. (Described by Zenkin et al., in the 28 July 2006 issue of Science.) |
Note that at any place in a DNA molecule, either strand may be serving as the template; that is, some genes "run" one way, some the other (and in a few remarkable cases, the same segment of double helix contains genetic information on both strands!). In all cases, however, RNA polymerase transcribes the DNA strand in its 3′ → 5′ direction.
| A report in the 4 January 2001 issue of Nature shows that RNA polymerase actually tracks around the double helix of DNA. In vitro, at least, when RNA polymerase is immobilized, it spins the DNA molecule around and around as it moves along the molecule. Whether it is the polymerase or the DNA that does the spinning in vivo remains to be determined. |
 | Sedimentation pattern produced by high-speed centrifugation of RNA extracted from the precursors of rabbit red blood cells. The discrete bands represent particular classes of RNA. The transfer RNAs band at about 4S. The ribosomal RNAs of eukaryotes sediment at 5S, 5.8S, 18S, and 28S. (The larger the sedimentation unit, S, the larger the molecule — but not proportionally.) The RNA forming the band at 9S is the messenger RNA for the synthesis of hemoglobin, the major protein synthesized in these cells. In most types of cells, the messenger RNAs are extremely heterogenous, with small amounts distributed from 6S to 25S. |
Messenger RNA will be translated into a polypeptide. Messenger RNA comes in a wide range of sizes reflecting the size of the polypeptide it encodes. Most cells produce small amounts of thousands of different mRNA molecules, each to be translated into a peptide needed by the cell.
Many mRNAs are common to most cells, encoding "housekeeping" proteins needed by all cells (e.g., the enzymes of glycolysis). Other mRNAs are specific for only certain types of cells. These encode proteins needed for the function of that particular cell (e.g., the mRNA for hemoglobin in the precursors of red blood cells).
The S number given each type of rRNA reflects the rate at which the molecules sediment in the ultracentrifuge. The larger the number, the larger the molecule (but not proportionally).
The 28S, 18S, and 5.8S molecules are produced by the processing of a single primary transcript from a cluster of identical copies of a single gene. The 5S molecules are produced from a different cluster of identical genes.
| Link to a view showing transcription in the rRNA gene cluster. |
These are the RNA molecules that carry amino acids to the growing polypeptide.
There are some 32 different kinds of tRNA in a typical eukaryotic cell. |  |
| Above: schematic showing the structure of alanine transfer RNA from yeast. The four helical regions formed by base pairing are shown. The loop on the left binds to the enzyme that catalyzes the attachment (by means of a "high-energy" bond) of alanine to the molecule (top). The anticodon loop carries the three nucleotides that recognize the appropriate codon on the messenger RNA. The circled letters indicate that the actual nucleotide is a chemically-modified form of the one shown. | Above: stereo view of phenylalanine transfer RNA from yeast. The 3' end where the phenylalanine is attached is at the upper right. The anticodon is located at the lower right. To fuse the two images, erect a stiff sheet of paper or cardboard between the two views so that your left eye sees only the left image and your right eye only the right image. With continued practice, you may even find that you can fuse the images with no props at all! (Courtesy of Dr. Sung-Hou Kim from J. L. Sussman and S-H. Kim, Science 192:853-858, May 28, 1976.) |
Approximately a dozen different genes for snRNAs, each present in multiple copies, have been identified. The snRNAs have various roles in the processing of the other classes of RNA. For example, several snRNAs are part of the spliceosomes that participate in converting pre-mRNA into mRNA by excising the introns and splicing the exons. [Link down to the discussion of RNA processing.]
In vertebrates, the snoRNAs are made from introns removed during RNA processing.
Only messenger RNA encodes polypeptides. All the other classes of RNA are thus called non-coding RNA. In addition to the rRNAs, snRNAs, and snoRNAs, there is a large (more than 10,000 in humans), heterogenous collection of transcripts longer than 200 nucleotides that are classified as lncRNAs. The function, if any, of most of these remains to be discovered.
However, some lncRNAs have been found to participate in the regulation of such diverse activities as
While much remains to be learned about their functions, taken together non-coding RNAs probably account for three-quarters of the transcription going on in the nucleus.
| Two remarkable reports in the 8 June 2001 issue of Science show the structure of Pol II and reveal many details of how it uses the antisense strand of DNA to synthesize a strand of mRNA. |
All the primary transcripts produced in the nucleus must undergo processing steps to produce functional RNA molecules for export to the cytosol. We shall confine ourselves to a view of the steps as they occur in the processing of pre-mRNA to mRNA.
Most eukaryotic genes are split into segments. In decoding the open reading frame of a gene for a known protein, one usually encounters periodic stretches of DNA calling for amino acids that do not occur in the actual protein product of that gene. Such stretches of DNA, which get transcribed into RNA but not translated into protein, are called introns. Those stretches of DNA that do code for amino acids in the protein are called exons. Examples:
In general, introns tend to be much longer than exons. An average eukaryotic exon is only 140 nucleotides long, but one human intron stretches for 480,000 nucleotides!
Removal of the introns — and splicing the exons together — are among the essential steps in synthesizing mRNA.
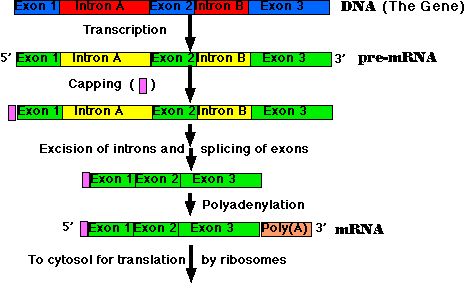
The steps of RNA processing:
The cutting and splicing of mRNA must be done with great precision. If even one nucleotide is left over from an intron or one is removed from an exon, the reading frame from that point on will be shifted, producing new codons specifying a totally different sequence of amino acids from that point to the end of the molecule (which often ends prematurely anyway when the shifted reading frame generates a STOP codon).
The removal of introns and splicing of exons is done by spliceosomes. These are a complexes of 5 snRNA molecules and some 145 different proteins.
The introns in most pre-mRNAs begin with a GU and end with an AG. Presumably these short sequences assist in guiding the spliceosome.

The upper image is an electron micrograph of a mRNA-DNA hybrid molecule formed by mixing the messenger RNA (mRNA) from a clone of antibody-secreting cells with single-stranded DNA from the same kind of cells. The bar represents the length of 1000 bases.
The lower diagram is an interpretation of the micrograph. The solid line represents the DNA; the dotted line the mRNA. The loops (IA, IB, etc.) represent the introns that separate the exons encoding the domains of an antibody heavy chain:The unhybridized portion of the mRNA is its poly(A) tail.
[From R. Maki et al., Proc. Natl. Acad. Sci. USA 77:2138, 1980.]
The processing of pre-mRNA for many proteins proceeds along various paths in different cells or under different conditions. For example, early in the differentiation of a B cell (a lymphocyte that synthesizes an antibody) the cell first uses an exon that encodes a transmembrane domain that causes the molecule to be retained at the cell surface. Later, the B cell switches to using a different exon whose domain enables the protein to be secreted from the cell as a circulating antibody molecule.
Alternative splicing provides a mechanism for producing a wide variety of proteins from a small number of genes. While we humans may turn out to have only some 20 thousand genes, we probably make at least 10 times that number of different proteins. It is now estimated that 92–94% of our genes produce pre-mRNAs that are alternatively-spliced. There is evidence that the pattern of alternative splicing differs consistently in different tissues and so must be regulated. But whether all the products are functional or that many are simply the outcome of an error-prone process remains to be seen.
Alternative splicing not only provides different proteins from a single gene but also different 3' UTRs and 5' UTRs. Although not translated into protein, these untranslated regions contain signals that, for example, dictate where in the cell that protein will accumulate. Two examples:One of the most dramatic examples of alternative splicing is the Dscam gene in Drosophila. This single gene contains some 116 exons of which 17 are retained in the final mRNA. Some exons are always included; others are selected from an array. Theoretically this system is able to produce 38,016 different proteins. And, in fact, over 18,000 different ones have been found in Drosophila hemolymph.
These Dscam proteins are used to establish a unique identity for each neuron. It works like this. Each developing neuron synthesizes a dozen or so Dscam mRNAs out of the thousands of possibilities. Which ones are selected appears to be simply a matter of chance, but because of the great number of possibilities, each neuron will most likely end up with a unique set of a dozen or so Dscam proteins. As each developing neuron in the central nervous system sprouts dendrites and an axon, these express its unique collection of Dscam proteins. If the various extensions of a single neuron should meet each other in the tangled web that is the hallmark of nervous tissue, they are repelled. In this way, thousands of different neurons can coexist in intimate contact without the danger of nonfunctional contacts between the various extensions of the same neuron.
We also have a DSCAM gene which is located on chromosome 21. When present in 3 copies, it is probably responsible for some of the features of Down syndrome (and accounts for its full name: Down Syndrome Cell Adhesion Molecule). However, it is not responsible for specifying neuron identity the way Dscam proteins do in Drosophila. That function appears to be accomplished by cell-surface proteins called protocadherins. Protocadherins are a subset of cadherins, proteins involved in cell-cell contact [Link]. Our protocadherins are encoded by a family of 53 genes. However, a single neuron expresses only about 6 of these. Which six are chosen occurs by chance alone, and this gives rise to thousands of possible combinations providing each neuron a unique identity. Like the process described above for Drosophila, if the dendrites of a single neuron should encounter each other, they avoid establishing a synapse by the repulsion mediated by their identical collection of protocadherins. In this way, thousands of different neurons can coexist in intimate contact without the danger of nonfunctional contacts between the various extensions of the same neuron. |
Whether a particular segment of RNA will be retained as an exon or excised as an intron can vary under different circumstances, such as
Most genes are transcribed and their transcripts processed as described above. RNA polymerase travels down a single strand of a single gene locus to form pre-mRNA that is processed (including removal of introns) to form the mature mRNA. But there are exceptions. A number of cases have been found where two different precursor transcripts have been spliced together to form the final RNA molecule. The phenomenon is called trans-splicing.
Examples: synthesis of a single RNA molecule by splicing together transcripts from lociPerhaps during evolution, eukaryotic genes have been assembled from smaller, primitive genes — today's exons. Some proteins, like the antibodies mentioned in the previous section, are organized in a set of separate sections or domains each with a special function to perform in the complete molecule. Each domain is encoded by a separate exon. Having the different functional parts of the antibody molecule encoded by separate exons makes it possible to use these units in different combinations. Thus a set of exons in the genome may be the genetic equivalent of the various modular pieces in a box of Lego® for children to assemble in whatever forms they wish.
But the boundaries of other exons do not seem to correspond to domain boundaries of the protein. So perhaps the major benefit of split genes is simply the opportunity they provide for making many different proteins from a single gene through alternative splicing.
In eukaryotes, transcription takes place in the nucleus, translation later on in the cytosol. In bacteria (they have no nucleus), both processes occur in the cytosol and simultaneously [View].
| Welcome&Next Search |