The human genome is presently estimated to contain 20–25 thousand genes. The number of T-cell receptors for antigen (TCRs) that we make is estimated at 2.5 x 107; the number of different kinds of antibody molecules (BCRs) is probably about the same.
How could 2.5 x 104 genes encode 2.5 x 107 different TCRs and the same number of different BCRs?
The answer: each receptor chain
- heavy (H) plus
kappa (κ) or lambda (λ) chains for BCRs;
- alpha (α) and beta (β) or gamma (γ) and delta (δ) chains for TCRs)
is encoded by several different gene segments.
The genome contains a pool of gene segments for each type of chain. Random assortment of these segments makes the largest contribution to receptor diversity.
For the heavy (H) chains of BCRs (antibodies), the gene segments are:
- 51 VH segments. Each of these encodes most of the N-terminal of the antibody, including the first two (but not the third) hypervariable or complementarity determining region (CDR).
- 27 DH (="diversity") gene segments. These encode part of the third CDR ("CDR3").
- 6 JH (="joining") gene segments. These encode the remainder of the V region of the BCR (including the remainder of CDR3).
- 9 CH gene segments. These encode the C region of the BCR (and the antibody derived from it). The C gene segments are
- 1 mu (µ); encodes the C region of IgM
- 1 delta (δ)
for IgD
- 4 gamma (γ) gene segments for the four types of IgG
- 1 epsilon (ε) for IgE
- 2 alpha (α) gene segments for the two types of IgA
All of these gene segments are clustered in a complex locus on chromosome 14.
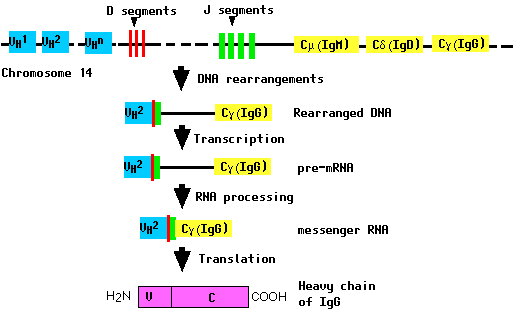
During the differentiation of the B cell (and long before any encounter with an antigen), the DNA in this locus is cut and recombined to make an intact gene for the heavy chain. This gene can then be transcribed into pre-mRNA, which is then processed to form the mRNA that will be translated into the heavy (H) polypeptide chain.
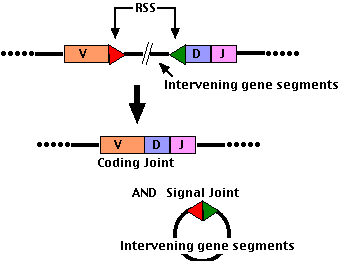
- Each gene segment (V, D, and J) has an adjacent Recombination Signal Sequence (RSS)
- at the 3' end of each V segment
- at both ends of each D segment
- at the 3' end of each J segment
- These are recognized by two proteins encoded by two Recombination Activating Genes
- The RAG-1 and RAG-2 proteins cut through both strands of DNA at the RSS forming double-stranded breaks (DSBs).
- Then the regular machinery for repairing DSBs (by nonhomologous end-joining) swings into action.
- The cut ends are stitched together (ligated) to form:
- a coding joint (D-J or V-DJ for heavy chains; V-J for light chains)
- a signal joint (usually a loop of DNA deleting all the intervening DNA initially present between the 2 gene segments chosen).
- D-J joining occurs first; then the combined DJ segment (still attached to the cluster of constant region gene segments) is joined to a V segment (as shown in the figure).
- The V gene segment chosen may be thousands of base pairs away from the D-J segment so the chromosome must be drawn into a loop to bring the two together.
The loop is stabilized by
- a protein designated CTCF ("CCCTC binding factor"; named for the nucleotide sequence to which it binds). The CTCF at the D-J site on the DNA forms a dimer with the CTCF at the V site on the DNA binding the two regions together.
- cohesin — the same protein complex that holds sister chromatids together during mitosis and meiosis. [Link]
- In the process, the cluster of gene segments moves from the periphery of the nucleus (a region of inactive genes) to the center of the nucleus (a region of active gene transcription). [Link]
Some cases of severe combined immunodeficiency in humans (SCID) are caused by defects in V(D)J joining.
- One version is caused by mutations in both copies of either RAG1 or RAG2.
- Another is caused by mutations in a gene needed for nonhomologous end-joining. (No coding joint is formed even though a signal joint forms normally.)
|
If the 51 VH, 27 DH, and 6 JH gene segments were assembled randomly (they probably are not), that would provide a minimum of 8.3 x 103 different possible combinations.
But the possibilities of antibody V region diversity turn out to be greater than that. The recombination process is not precise.
- the exact points of splicing
- between VH and DH and
- between DH and JH
can vary over several nucleotides
- extra nucleotides, called N regions, can also be inserted at these joints.
- All of these add greatly to the diversity of CDR3.
Light chains
Once the H chain gene is assembled, transcribed, and translated, the resulting H chain can pair with an L chain that is itself the product of a similar recombination process occurring
- on chromosome 2 for kappa gene segments;
- on chromosome 22 for lambda gene segments.
| Antibodies (BCRs) | Gene Segments | Combinations |
|---|
| Vκ |
40 | |
| Jκ | 5 | 200 κ chains |
| Vλ | 31 | |
| Jλ | 4 | 124 λ chains |
| VH | 51 | |
| DH | 25 | |
| JH | 6 | 7,650 H chains |
| Any H chain with any L chain (324) | 2.5 x 106 |
As the table shows, this lays the foundation for a potential B-cell repertoire of 2.5 x 106 different antibody V regions. But the true number is probably virtually limitless because of
- variation in the exact splice point and
- the introduction of N nucleotides
- both of which increase the diversity of CDR3.
The combining of V, D, and J gene segments coupled with the random incorporation of extra nucleotides (N regions) at the joints, creates enormous coding variability. It also creates a high risk (two times out of three) of introducing a frameshift so that the codons for the rest of the V region encode nonsense. Although many B cells are wasted, the odds are not quite as bad as they seem:
- If the B cell fails to make a functional product from the cluster of gene segments on one of its chromosomes, it can turn to the gene segments on its homolog and try again.
- If it fails both times to make a functional kappa L chains, it still has two tries at making a functional lambda L chain.
Somatic Hypermutation (SHM) and Antibody Diversity
The diversifying mechanisms described above take place before the B cell encounters antigen. After a B cell encounters antigen, it may begin mitosis, growing into a clone of cells synthesizing the same BCR (and, eventually, secreting antibodies with the same binding site). Point mutations can occur while this is going on. Some of these may generate a binding site with increased affinity for its epitope. These are favorable mutations, and the "subclone" in which they occur tends to be favored and may replace the ancestral clone. The result is affinity maturation — the production of antibodies of ever-increasing affinity for the antigen.
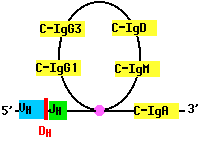
As B cells grow into a clone in response to antigen, they may rearrange their DNA once again. For example, a B cell that has assembled a complete gene for the H chain of IgM (µ), may cut the gene on the 3´ side of the assembled V-region segments and move the assembly to the 5´ side of another of its CH gene segments. Now the cell begins to make a different class of antibody, such as IgG or IgA. But the antigen specificity of the antibody remains the same because the N-terminal of the H chain remains unchanged (as does the entire L chain).
Class switch recombination enables the body to produce antibodies with different effector functions; that is, different means of dealing with the same antigen.
The ability of a B cell to switch CH gene segments depends on its receiving help from helper T cells.
The most abundant T cells in the blood express a receptor for antigen (TCR) that is a heterodimer of two chains designated alpha (α) and beta (β).
Each of these is encoded by a gene assembled from V, D, J, and C gene segments. Like BCRs, there are multiple variants of these gene segments arranged in clusters:
- alpha chain gene segments on chromosome 14 and
- beta chain gene segments on chromosome 7
| T cell receptors (TCRs) | Gene segments | Combinations |
|---|
| Vα | 50 | |
| Jα | 50 | 2.5 x 103 alpha chains |
| Vβ | 20 | |
| Jβ | 13 | |
| Dβ | 2 | 520 beta chains |
| Any alpha with any beta chain | 1.3 x 106 |
And like B cells, the greatest diversity in the receptors of αβ T cells occurs in the third complementarity determining region (CDR3) of the alpha and beta chains because of
- the junctional diversity between the V, J, and D segments and
- the addition of N region nucleotides.
However, T cells do not seem to use somatic mutation to increase receptor diversity. Actual measurements of the repertoire in humans reveals a figure of about 2.5 x 107.
The TCR repertoire of γδ T cells seems much smaller than that of their αβ cousins. The - gamma chain gene segments are clustered on chromosome 7;
- the delta chain gene segments are clustered on chromosome 14 (within the alpha chain cluster)
22 August 2015


