Transposons are segments of DNA that can move around to different positions in the genome of a single cell. In the process, they may
- cause mutations
- increase (or decrease) the amount of DNA in the genome of the cell, and if the cell is the precursor of a gamete, in the genomes of any descendants.
These mobile segments of DNA are sometimes called "jumping genes".
There are two distinct types:
- Class II transposons. These consist of DNA that moves directly from place to place.
- Class I transposons. These are retrotransposons that
- first transcribe the DNA into RNA and then
- use reverse transcriptase to make a DNA copy of the RNA to insert in a new location.
Class II transposons move by a "cut and paste" process: the transposon is cut out of its location (like command/control-X on your computer) and inserted into a new location (command/control-V).
This process requires an enzyme — a transposase — that is encoded within some of these transposons.
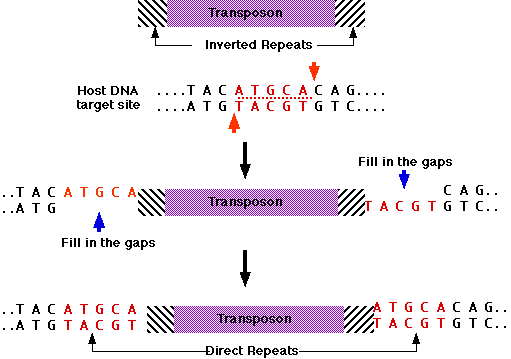 Transposase binds to:
Transposase binds to:
- both ends of the transposon, which consist of inverted repeats; that is, identical sequences reading in opposite directions.
- a sequence of DNA that makes up the target site. Some transposases require a specific sequence as their target site; others can insert the transposon anywhere in the genome.
The DNA at the target site is cut in an offset manner (like the "sticky ends" produced by some restriction enzymes [Examples]).
After the transposon is ligated to the host DNA, the gaps are filled in by Watson-Crick base pairing. This creates identical direct repeats at each end of the transposon.
Often transposons lose their gene for transposase. But as long as somewhere in the cell there is a transposon that can synthesize the enzyme, their inverted repeats are recognized and they, too, can be moved to a new location.
The recent completion of the genome sequence of rice and C. elegans has revealed that their genomes contain thousands of copies of a recurring motif consisting of
- almost identical sequences of about 400 base pairs flanked by
- characteristic inverted repeats of about 15 base pairs such as
5' GGCCAGTCACAATGG..~400 nt..CCATTGTGACTGGCC 3'
3' CCGGTCAGTGTTACC..~400 nt..GGTAACACTGACCGG 5'
MITEs are too small to encode any protein. Just how they are copied and moved to new locations is still uncertain. Probably larger transposons that
- do encode the necessary enzyme and
- recognize the same inverted repeats
are responsible.
There are over 100,000 MITEs in the rice genome (representing some 6% of the total genome). Some of the mutations found in certain strains of rice are caused by the insertion of a MITE in the gene.
MITEs have also been found in the genomes of humans, Xenopus, and apples.
The first transposons were discovered in the 1940s by Barbara McClintock who worked with maize (Zea mays, called "corn" in the U.S.). She found that they were responsible for a variety of types of gene mutations, usually

| Some of the mutations (c, bz) used as examples of how gene loci are mapped on the chromosome were caused by transposons. [Link] |
In developing somatic tissues like corn kernels, a mutation (e.g., c) that alters color will be passed on to all the descendant cells. This produces the variegated pattern which is so prized in "Indian corn". (Photo courtesy of Whalls Farms.)
It took about 40 years for other scientists to fully appreciate the significance of Barbara McClintock's discoveries. She was finally awarded a Nobel Prize in 1983.
P elements are Class II transposons found in Drosophila. They do little harm because expression of their transposase gene is usually repressed. However, when male flies with P elements mate with female flies lacking them, the transposase becomes active in the germline producing so many mutations that their offspring are sterile.
In nature this is no longer a problem. P elements seem to have first appeared in Drosophila melanogaster about 50 years ago. Since then, they have spread through every population of the species. Today flies lacking P elements can only be found in old strains maintained in the laboratory.
P elements have provided valuable tools for Drosophila geneticists. Transgenic flies containing any desired gene can be produced by injecting the early embryo with an engineered P element containing that gene.
Other transposons are being studied for their ability to create transgenic insects of agricultural and public health importance.
Some transposons in bacteria carry — in addition to the gene for transposase — genes for one or more (usually more) proteins imparting resistance to antibiotics. When such a transposon is incorporated in a plasmid, it can leave the host cell and move to another. This is the way that the alarming phenomenon of multidrug antibiotic resistance spreads so rapidly.
Transposition in these cases occurs by a "copy and paste" (command/control-C -> command/control-V) mechanism. This requires an additional enzyme — a resolvase — that is also encoded in the transposon itself. The original transposon remains at the original site while its copy is inserted at a new site.
Retrotransposons also move by a "copy and paste" mechanism but in contrast to the transposons described above, the copy is made of RNA, not DNA.
The RNA copies are then transcribed back into DNA — using a reverse transcriptase — and these are inserted into new locations in the genome.
Many retrotransposons have long terminal repeats (LTRs) at their ends that may contain over 1000 base pairs in each.
Like DNA transposons, retrotransposons generate direct repeats at their new sites of insertion. In fact, it is the presence of these direct repeats that often is the clue that the intervening stretch of DNA arrived there by retrotransposition.
Some 40% of the entire human genome consists of retrotransposons.
- The human genome contains some 500,000 LINEs (representing some 17% of the genome).
- The most abundant of these belong to a family called LINE-1 (L1).
- These L1 elements are DNA sequences that range in length from a few hundred to as many as 9,000 base pairs.
- Only about 50 L1 elements are functional "genes"; that is, can be transcribed and translated.
- The functional L1 elements are about 6,500 bp in length and encode three proteins, including
- an endonuclease that cuts DNA and a
- reverse transcriptase that makes a DNA copy of an RNA transcript.
- L1 activity proceeds as follows:
- RNA polymerase II transcribes the L1 DNA into RNA.
- The RNA is translated by ribosomes in the cytoplasm into the proteins.
- The proteins and RNA join together and reenter the nucleus.
- The endonuclease cuts a strand of "target" DNA, often in the intron of a gene.
- The reverse transcriptase copies the L1 RNA into L1 DNA which is inserted into the target DNA forming a new L1 element there.
Through this copy-paste mechanism, the number of LINEs can increase in the genome.
The oncogene p53 suppresses the formation of new L1 elements. If p53 becomes non-functional because of mutation, as is the case in many cancers, the number of L1 elements increases. Occasionly, insertion of a new L1 element breaks the chromosome. Such breaks are common in cancer cells.
The diversity of LINEs between individual human genomes make them useful markers for DNA "fingerprinting".
Variation occurs in the length of L1 elements:
- Transcription of an active L1 element sometimes continues downstream into additional DNA producing a longer transposed element.
- Reverse transcription of L1 RNA often concludes prematurely and produces a shortened transposed element.
While most L1 elements are not functional, they may play a role in regulating the efficiency of transcription of the gene in which they reside (see below).
Occasionally, L1 activity makes and inserts a copy of a cellular mRNA (thus a natural cDNA). Lacking introns as well as the necessary control elements like promoters, these genes are not expressed. They represent one category of pseudogene.
SINEs are short DNA sequences (100–400 base pairs) that represent reverse-transcribed RNA molecules originally transcribed by RNA polymerase III; that is, molecules of tRNA, 5S rRNA, and some other small nuclear RNAs.
There are around 1.8 million copies in the human genome (representing some 10% of our total DNA).
The most abundant SINEs are the Alu elements. Alu elements consist of a sequence averaging 260 base pairs that contains a site that is recognized by the restriction enzyme AluI. They appear to be reverse transcripts of 7S RNA, part of the signal recognition particle.
Most SINEs do not encode any functional molecules and depend on the machinery of active L1 elements to be transposed; that is, copied and pasted in new locations.
HIV-1 — the cause of AIDS — and other human retroviruses (e.g., HTLV-1, the human T-cell leukemia/lymphoma virus) behave like retrotransposons.
The RNA genome of HIV-1 contains a gene for
- reverse transcriptase and one for
- integrase. The integrase serves the same function as the transposases of DNA transposons. The DNA copies can be inserted anywhere in the genome.
Molecules of both enzymes are incorporated in the virus particle.
Transposons are mutagens. They can cause mutations in several ways:
- If a transposon inserts itself into a functional gene, it will probably damage it. Insertion into exons, introns, and even into DNA flanking the genes (which may contain promoters and enhancers) can destroy or alter the gene's activity.
| The insertion of a retrotransposon in the DNA flanking a gene for pigment synthesis is thought to have produced white grapes from a black-skinned ancestor. Later, the loss of that retrotransposon produced the red-skinned grape varieties cultivated today. |
- Faulty repair of the gap left at the old site (in cut and paste transposition) can lead to mutation there.
- The presence of a string of identical repeated sequences presents a problem for precise pairing during meiosis. How is the third, say, of a string of five Alu sequences on the "invading strand" of one chromatid going to ensure that it pairs with the third sequence in the other strand? If it accidentally pairs with one of the other Alu sequences, the result will be an unequal crossover — one of the commonest causes of duplications.
SINEs (mostly Alu sequences) and LINEs cause only a small percentage of human mutations. (There may even be a mechanism by which they avoid inserting themselves into functional genes.) However, they have been found to be the cause of the mutations responsible for some cases of human genetic diseases, including:
Transposons have been called "junk" DNA and "selfish" DNA.
- "selfish" because their only function seems to make more copies of themselves and
- "junk" because there is no obvious benefit to their host.
Because of the sequence similarities of all the LINEs and SINEs, they also make up a large portion of the "repetitive DNA" of the cell.
Retrotransposons cannot be so selfish that they reduce the survival of their host. And it now appears that many, at least, confer some benefit. The ENCODE project found that some 75% of our repetitive DNA occurs within, or overlaps with, sequences, like enhancers, that regulate gene expression.
Some other possibilities:
- Retrotransposons often carry some additional sequences at their 3' end as they insert into a new location. Perhaps these occasionally create new combinations of exons, promoters, and enhancers that benefit the host.
Example:
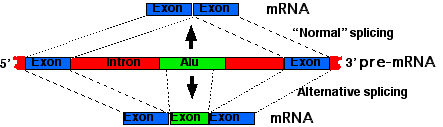
- Thousands of our Alu elements occur in the introns of genes.
- Some of these contain sequences that when transcribed into the primary transcript are recognized by the spliceosome.
- These can then be spliced into the mature mRNA creating a
- new exon, which will be transcribed into a new protein product.
- Alternative splicing can provide not only the new mRNA (and thus protein) but also the old.
- In this way, nature can try out new proteins without the risk of abandoning the tried-and-true old one.
- L1 elements inserted into the introns of functional genes reduce the transcription of those genes without harming the gene product — the longer the L1 element, the lower the level of gene expression. Some 79% of our genes contain L1 elements, and perhaps they are a mechanism for establishing the baseline level of gene activity.
- Telomerase, the enzyme essential for maintaining chromosome length, is closely related to the reverse transcriptase of LINEs and may have evolved from it.
- RAG-1 and RAG-2. The proteins encoded by these genes are needed to assemble the repertoire of antibodies and T-cells receptors (TCRs) used by the adaptive immune system [Link]. The mechanism [Link] resembles that of the cut and paste method of Class II transposons , and the RAG genes may have evolved from them. If so, the event occurred some 450 million years ago when the jawed vertebrates evolved from jawless ancestors [Link]. Only jawed vertebrates have the RAG-1 and RAG-2 genes.
- In Drosophila, the insertion of transposons into genes has been linked to the development of resistance to DDT and organophosphate insecticides.
- The genome of Arabidopsis thaliana contains ~1.2 x 108 base pairs (bp) of DNA. About 14% of this consists of transposons; the rest functional genes (25,498 of them).
- The maize (corn) genome contains 20 times more DNA (2.4 x 109 bp) but surely has no need for 20 times as many genes. In fact, 60% of the corn genome is made up of transposons. (The figure for humans is 42%.)
So it seems likely that the lack of an association between size of genome and number of functional genes — the C-value paradox — is caused by the amount of transposon DNA accumulated in the genome.
9 March 2024

 Transposase binds to:
Transposase binds to:
